
Cells package their DNA not only to protect it, but also to regulate which genes are accessed and when. Cellular genes are therefore similar to valuable files stored in a file cabinet — but in this case, the cabinet's drawers are constantly opening and closing; various files are continually being located, pulled, and copied; and the original files are always returned to the correct location.
Of course, just as file drawers help conserve space in an office, DNA packaging helps conserve space in cells. Packaging is the reason why the approximately two meters of human DNA can fit into a cell that is only a few micrometers wide. But how, exactly, is DNA compacted to fit within eukaryotic and prokaryotic cells? And what mechanisms do cells use to access this highly compacted genetic material?
Cellular DNA is never bare and unaccompanied by other proteins. Rather, it always forms a complex with various protein partners that help package it into such a tiny space. This DNA-protein complex is called chromatin, wherein the mass of protein and nucleic acid is nearly equal. Within cells, chromatin usually folds into characteristic formations called chromosomes. Each chromosome contains a single double-stranded piece of DNA along with the aforementioned packaging proteins.

During interphase (1), chromatin is in its least condensed state and appears loosely distributed throughout the nucleus. Chromatin condensation begins during prophase (2) and chromosomes become visible. Chromosomes remain condensed throughout the various stages of mitosis (2-5).
![]()
© 2013 Nature Education All rights reserved.
Eukaryotes typically possess multiple pairs of linear chromosomes, all of which are contained in the cellular nucleus, and these chromosomes have characteristic and changeable forms. During cell division, for example, they become more tightly packed, and their condensed form can be visualized with a light microscope. This condensed form is approximately 10,000 times shorter than the linear DNA strand would be if it was devoid of proteins and pulled taut. However, when eukaryotic cells are not dividing — a stage called interphase — the chromatin within their chromosomes is less tightly packed. This looser configuration is important because it permits transcription to take place (Figure 1, Figure 2).
In contrast to eukaryotes, the DNA in prokaryotic cells is generally present in a single circular chromosome that is located in the cytoplasm. (Recall that prokaryotic cells do not possess a nucleus.) Prokaryotic chromosomes are less condensed than their eukaryotic counterparts and don't have easily identified features when viewed under a light microscope.
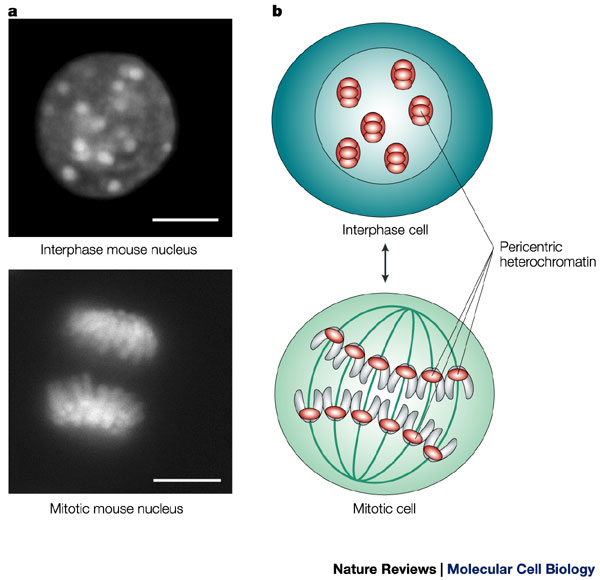
During interphase, the cell's DNA is not condensed and is loosely distributed. A stain for heterochromatin (which indicates the position of chromosomes) shows this broad distribution of chromatin in a mouse cell (upper left). The same stain also shows the organized, aligned structure of the chromosomes during mitosis. Scale bars = 10 microns.
![]()
© 2004 Nature Publishing Group Maison, C. & Almouzni, G. HP1 and the dynamics of heterochromatin maintenance. Nature Reviews Molecular Cell Biology 5, 296-305 (2004). All rights reserved.

Eukaryotic chromosomes consist of repeated units of chromatin called nucleosomes, which were discovered by chemically digesting cellular nuclei and stripping away as much of the outer protein packaging from the DNA as possible. The chromatin that resisted digestion had the appearance of "beads on a string" in electron micrographs — with the "beads" being nucleosomes positioned at intervals along the length of the DNA molecule (Figure 3).
Nucleosomes are made up of double-stranded DNA that has complexed with small proteins called histones. The core particle of each nucleosome consists of eight histone molecules, two each of four different histone types: H2A, H2B, H3, and H4. The structure of histones has been strongly conserved across evolution, suggesting that their DNA packaging function is crucially important to all eukaryotic cells (Figure 4).
Histones carry positive charges and bind negatively charged DNA in a specific conformation. In particular, a segment of the DNA double helix wraps around each histone core particle a little less than twice. The exact length of the DNA segment associated with each histone core varies from species to species, but most such segments are approximately 150 base pairs in length. Furthermore, each histone molecule within the core particle has one end that sticks out from the particle. These ends are called N-terminal tails, and they play an important role in higher-order chromatin structure and gene expression.

Each nucleosome contains eight histone proteins (blue), and DNA wraps around these histone structures to achieve a more condensed coiled form.
![]()
© 2010 Nature Education All rights reserved.